Prokaryotic diversity changes and their functional interrelationship with land use
Acidobacteria form a widespread and abundant but little studied phylum of bacteria. Based on molecular studies of the 16S rRNA genes, Acidobacteria have been detected in various ecosystems such as soils and sediments, hot springs, peat bogs, plankton of the deep Mediterranean Sea, acidic lakes of spoil heaps and caves.
Acidobacteria can account for up to 50 to 80% of soil bacteria. The phylogenetic diversity of Acidobacteria and their abundance in soil is thought to be similar to that of the phylum Proteobacteria. This phylogenetic diversity and the wide distribution and abundance especially in soil habitats indicate an important role in biogeochemical processes and a high metabolic versatility of these bacteria. However, almost nothing is known about the functional role of Acidobacteria and the connection to land use.
1. Acidobacteria are a dominant group of soil bacteria and relevant for ecosystem functions in soils.
2. The diversity and activity of Acidobacteria correlate with plant diversity. 3. land use is a factor influencing the functional diversity of Acidobacteria.
3. Land use influences the functional diversity of Acidobacteria.
In the present project, the functional relevance of diversity changes is to be investigated using a land use gradient as the main variable. The objectives are to record the composition, the key physiological functions and the identification of important functional groups of Acidobacteria.
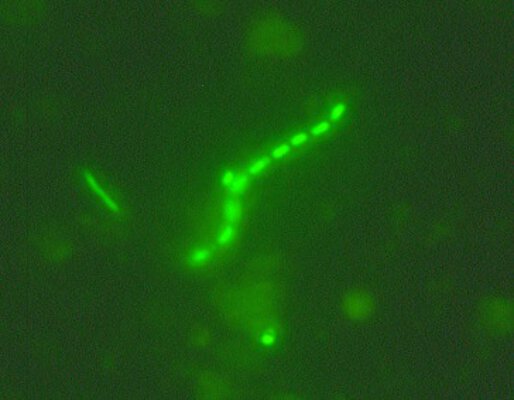
Diversity changes and physiologically active phylotypes are investigated using so-called fingerprinting methods, DGGE and T-RFLP. To identify dominant populations, 16S rRNA clone libraries are created and corresponding clones are sequenced. To determine physiological properties, existing metagenome libraries will be used and representative representatives will be isolated using novel cultivation methods. By adding different 13C carbon sources and following the incorporation of these substrates by stable isotope probing of RNA, we aim to elucidate the functional role of some important phylotypes. Furthermore, we investigate the seasonal variation of Acidobacteria communities by cell counts, DGGE and T-RFLP analysis and sample all 57 VIPs in April, June, August and October 2009.
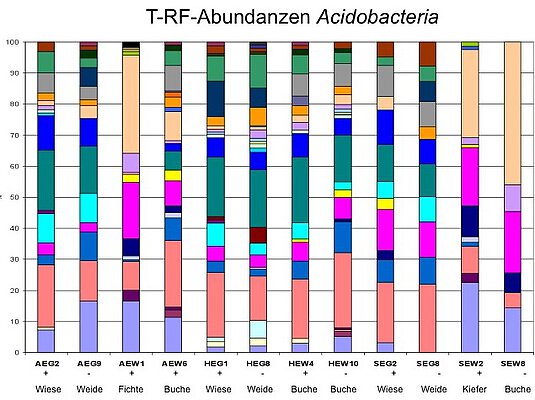
Cell counts so far showed that the Schwäbische Alb exploratory has the most cells per g of soil, followed by Hainich-Dün and Schorfheide-Chorin. Cluster analyses of the acidobacterial communities recorded by DGGE and T-RFLP analyses showed that soil samples from an exploratory generally cluster together but are separated by the type of land use (forest/grassland). The uneven distribution and abundance of acidobacteria in forest and grassland thus leads to separate clusters. Especially the forest soils of the Schorfheide differ significantly from all other soils and form their own cluster. The occurrence of Acidobacteria is strongly pH-dependent; consequently, these results probably result from different pH values in forest (~ pH 5.0) and grassland (~ pH 6.5), whereby the forest soil in the Schorfheide-Chorin exploratory showed by far the lowest pH values (~ pH 3.5).