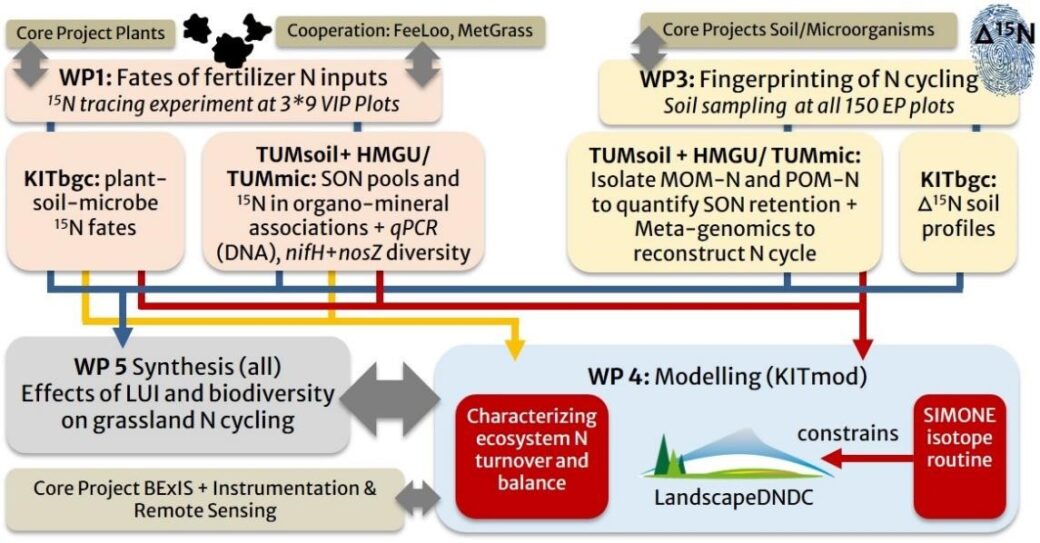
Linking land use intensity, biodiversity, soil microbial processes and organo-mineral interactions for a mechanistic understanding of nitrogen (N) turnover in grassland ecosystems
Increasing land use intensity to produce more protein-rich fodder might be directly linked with detrimental effects on biodiversity and multifunctionality of grassland ecosystems and drive environmental nitrogen losses. Still, interactions between land use intensity, above- and belowground biodiversity and nitrogen (N) cycling in grassland ecosystems lack a mechanistic understanding, which prevents targeted mitigation measures. Through an interdisciplinary integration of soil microbial ecology, short-term processes of biotic N partitioning in the plant-soil-microbe system, and long-term physicochemical soil organic N retention pathways, BE_BioMon aims to fill this knowledge gap by linking measurements and modelling.
With this holistic measurement and modelling work we expect to contribute to an improved mechanistic and functional understanding of interactions of land use intensity, biodiversity and grassland ecosystem N cycling as a basis for mitigating environmental impacts. As a new project, we openly look forward to collaborative opportunities and the further exchange of ideas with all Explorers.
Our overarching goal is to improve the mechanistic understanding of interactions between land use intensity and biodiversity as a driver of grassland N turnover. More specifically, our objectives are:
O1: To assess effects of land use intensity and above- and belowground biodiversity on biotic and abiotic processes of fertilizer N partitioning in the plant-soil-microbe system of grassland ecosystems.
O2: To fingerprint land use intensity effects on grassland N cycling combining vertical δ15N soil profiles, functional microbiome fingerprinting (metagenomics), and soil organic N fractionation approaches.
O3: To synthesize project measurements and existing data of the Biodiversity Exploratories for further development and advanced testing of a process based biogeochemical model.
Our overarching hypothesis is that effects of land use intensity on ecosystem N cycling processes are closely linked to above- and belowground biodiversity, with highest biodiversity and fastest utilization and turnover of N inputs under intermediate land use intensity, and only small modulation by site-specific conditions. More specifically, we hypothesize:
H1: With decreasing land use intensity, increasing C:N ratio and diversity of heterotrophic microorganisms, microbial N retention of fertilizer N inputs increases because of a shift from nitrification to inorganic N immobilization. Such N assimilation by a highly diverse microbiome promotes formation of POM- and MOM-N through microbial necromass that is affected by soil aggregate turnover.
H2: Soil δ15N increases with increasing land use intensity due to higher manure inputs, higher fractionation through increased rates of nitrification and denitrification and decreasing abundance, diversity and activity of diazotrophs. Consequently, it has a high potential to be used as a process integrating benchmark quantity for ecosystem models.
H3: The measurements of microbial diversity/ activity, spatiotemporal dynamics of N pools and fluxes as well as isotopic signatures will allow for improved process description, parameterization and validation of a process-based model. The improved model is an essential tool for spatiotemporal scaling and to assess impacts of land use intensity and microbial/ plant diversity on N cycling and losses across Biodiversity Exploratories grassland plots.
In this project, we follow the fate of organic fertilizer or feces-derived N in the plant-soil-microbe system using 15N stable isotope tracing approaches at selected plots of all three Biodiversity Exploratories. Detailed process understanding linked to the soil microbiome and aggregate structure as derived from these field experiments will advance the ecosystem model Landscape-DNDC to facilitate modelling of land use intensity effects on grassland ecosystem N cycling at plot to landscape scales. In this context, the natural 15N isotope “fingerprints” in soil profiles down to 1 m obtained during the soil sampling campaign in 2023 will serve as independent benchmark data for model validation.